1. Introduction
The OrganismTagger is a hybrid rule-based/machine-learning system that extracts organism mentions from the biomedical literature, normalizes them to their scientific name, and provides grounding to the NCBI Taxonomy database. Our pipeline provides the flexibility of annotating the species of particular interest to bio-engineers on different corpora, by optionally including detection of common names, acronyms, and strains. The OrganismTagger performance has been evaluated on two manually annotated corpora, OT and Linneaus [9]. On the OT corpus, the OrganismTagger achieves a precision and recall of 95% and 94% and a grounding accuracy of 97.5%. On the manually annotated corpus of Linneaus-100, the results show a precision and recall of 99% and 97% and grounding with an accuracy of 97.4%. It is published as open source software and described in detail in our publication, "OrganismTagger: Detection, normalization, and grounding of organism entities in biomedical documents [14]", Bioinformatics, vol. 27, no. 19 Oxford University Press, pp. 2721--2729, August 9, 2011.
2. Features
The OrganismTagger has a number of advanced features for species name recognition (for the details, please read the documentation and our paper):
- Named Entity Detection: each occurrence of an organism (species name) is semantically annotated as an Organism, with additional features showing the genus and species parts.
- Normalization: organism names can also be abbreviated or appear as acronyms, both of which are recognized as well. Additionally, each detected mention is normalized by adding the full, scientific name as a feature. The OrganismTagger includes a novel normalization heuristic that can find the full form even if it does not appear in the document.
- Grounding: each recognized and normalized mention additionally receives an NcbiId feature that links to its entry in the NCBI Taxonomy Database [16] — this is especially useful for disambiguating other biological entities in a document. An additional url feature directly links to the organism's web page on NCBI Taxonomy.
- Strain Recognition: using a novel machine learning approach, the OrganismTagger also detects and annotates strain-level information.
- Updateability: the OrganismTagger resources (gazetteering lists, RDF files, etc.) are automatically generated from a download of the NCBI database. All scripts are included to allow end users to update (and customize) their own installation.
- Web Service: you can run the OrganismTagger as a Web service, using standard SOAP requests; an OWL service description for the Semantic Assistants architecture [17] is included with the distribution. We also provide a demo Web service in case you want to try it out before running your own installation.
- Desktop Clients: the Semantic Assistants also allow you to run the OrganismTagger interactively using a number of desktop plug-ins, like for Firefox or OpenOffice.
- Training Data and Corpora: the distribution includes our manual annotations for evaluation as well as the training data and configuration files for the machine learning algorithm in case you want to experiment with improving the performance.
- Customization: the OrganismTagger is highly customizable, for details on run-time parameters please refer to the user's guide. The default configuration, as included in the demo pipeline, provides generally excellent performance.
- Scalability: based on the General Architecture for Text Engineering [18] (GATE), you can easily batch-process large amounts of documents by deploying the OrganismTagger with the GATECloud Paralleliser. On a standard quad-core desktop processor, OrganismTagger can process roughly 7 full-text papers/second (i.e., 100 full-text papers in ~14 seconds).
 [15]OrganismTagger annotations displayed in Firefox
[15]OrganismTagger annotations displayed in Firefox
3. Result Annotations
 OrganismTagger annotation displayed in GATE Developer
OrganismTagger annotation displayed in GATE Developer
The output of the OrganismTagger is a semantic annotation of the textual entities representing organisms, including the detected information from normalization and grounding. The screen shot shows the organism P. fluorescens subsp. cellulosa with its annotations displayed in the GATE [19] Developer GUI. The genus, species and subspecies parts, as well as the scientific name are based on the NCBI Taxonomy database [16]. You can also see the normalization of the abbreviation and the grounding of the detected organism to the NCBI Taxonomy database. NCBI IDs and the scientific names for both the taxa level and full form of the organism are provided. In detail, the features of an organism annotation are:
- Found: This feature is added through normalization: If the search heuristic based on the non-abbreviated form in the document is successful, this feature is set to FullName, indicating that the non-abbreviated form is available in the document. If the first heuristic fails, and the abbreviated mention is resolved through the second heuristic, it is set to Genus.
- Genus: Gives the genus part of the organism as it appears in the text (either the abbreviated form or the non-abbreviated one).
- Rule: Specifies which particular rule detected the organism mention.
- Species: Denotes the species name of the organism.
- Subspecies: In case the subspecies part of the organism is mentioned, it is detected and added to the feature list.
- Strain: Indicates the detected strain part of the organism.
- TaxalevelID: For the organism mentions with a more precise subspecies or strain level identification, the NCBI ID for the taxa level is also provided by the TaxalevelID feature during the grounding step.
- TaxalevelscientificName: The scientific name of the taxa level, as provided by the NCBI Taxonomy database.
- abbrGenus: In case the abbreviated form of the genus is used, this feature is set to true.
- class: The ontology class of the detected entity, here Organism.
- docName: The matched text in the document.
- instanceName: The standardized name that can be used for ontology population.
- ncbiId: The corresponding NCBI ID for the organism mention.
- organismName: The standard name as given by the NCBI Taxonomy database.
- scientificName: This feature provides the scientific name of the organism.
- url: A hyperlink connecting uniquely grounded organisms to their entry in the NCBI Taxonomy database.
4. Corpora
Tags from two manually annotated corpora, (i) a corpus of documents on protein engineering and fungi (ii) a corpus of open access biomedical documents from the PMC from the Linnaeus [9] project are also provided. The organism mention tags are in tab-delimited text files:
| NCBI ID | Document | Start | End | Text |
| 51453 | 15260499 | 754 | 772 | Trichoderma reesei |
| 51453 | 15260499 | 9295 | 9304 | T. reesei |
| 51453 | 15260499 | 9867 | 9891 | T. reesei Rut-C30 strain | 562 | 15260499 | 9999 | 10015 | Escherichia coli |
5. Quick Start Guide
As our OrganismTagger comes in form of a GATE [19] pipeline, you will need GATE itself. Note that you will need GATE version 7.0 or better to run the OrganismTagger. You can install the system directly from within GATE using the CREOLE Plugin Manager by selecting our Semantic Software Lab repository. Please note that you will need a larger than default memory setting to run the OrganismTagger, we recommend 1.5GB RAM per thread (2GB RAM per thread when using 64bit Java). Under Linux, you can start GATE in the following way: bin/gate.sh -Xmx1500M.
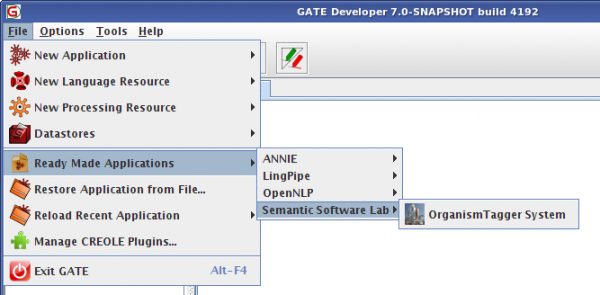 [20]OrganismTagger System in GATE Developer
[20]OrganismTagger System in GATE Developer
6. Running the OrganismTagger as a Web Service
The OrganismTagger can also be used as a Web service, either by itself or when integrated into a more complex pipeline. Towards this end, an OWL service description for the Semantic Assistants [21] framework is included in the release distribution. Simply follow these two steps, and after re-loading the SA server, it will now offer an "OrganismTagger" service:
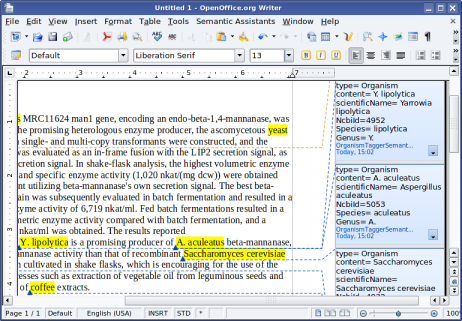 [22]Text abstract shown from Heterologous expression and optimized production of an Aspergillus aculeatus endo-1,4-beta-mannanase in Yarrowia lipolytica, PMID: 19507068, processed by the OrganismTagger running in OpenOffice
[22]Text abstract shown from Heterologous expression and optimized production of an Aspergillus aculeatus endo-1,4-beta-mannanase in Yarrowia lipolytica, PMID: 19507068, processed by the OrganismTagger running in OpenOffice
- Copy the organismtagger.owl service description into the directory Resources/OwlServiceDescriptions/ inside your semantic-assist installation
- Copy the directory "gate" to Resources/GatePipelines and rename it to "OrganismTagger"
Either by using the command line client that ships with the Semantic Assistants architecture [23] or any other Semantic Assistants-enabled client, like OpenOffice [24], you can execute the service and process input documents. Of course, the server also accepts plain SOAP requests. Our server minion.cs.concordia.ca (port 8879) usually offers the "OrganismTagger" service, if you want to test it.
7. Download
Latest version is v1.5 from 24.09.2014. You can install this version directly from within GATE through the CREOLE Plugin Manager. The download includes documentation, the example pipeline, and our annotated corpora. You can download the install package [25] manually (but the recommended way of installation is to use the GATE plugin manager through the GATE Developer GUI).
You can also look at the documentation [26] (this is the same file as included in the distribution in the doc/ folder).